Python image processing libraries performance: OpenCV vs Scipy vs Scikit-Image
image-processing numpy opencv python scikit-image scipyWe are going to compare the performance of different methods of image processing using three Python libraries (scipy, opencv and scikit-image). All the tests will be done using timeit. Also, in the case of OpenCV the tests will be done with the optimization enabled (by default if supported by CPU) and disabled:
>>> cv2.useOptimized() True >>> cv2.setUseOptimized(False) >>> cv2.useOptimized() False
Requirements
% sudo apt-get install -y build-essential libblas-dev liblapack-dev gfortran cmake git \ libgtk2.0-dev pkg-config libavcodec-dev libavformat-dev libswscale-dev python-dev \ python-numpy libtbb2 libtbb-dev libjpeg-dev libpng-dev libtiff-dev libjasper-dev libdc1394-22-dev % sudo pip install scipy scikit-image % mkdir tmp && cd tmp % git clone https://github.com/Itseez/opencv.git % git clone https://github.com/Itseez/opencv_contrib.git % cd opencv % mkdir build && cd build % cmake -D CMAKE_BUILD_TYPE=Release -D CMAKE_INSTALL_PREFIX=/usr/local .. % make -j7 % sudo make install % bin/opencv_test_core
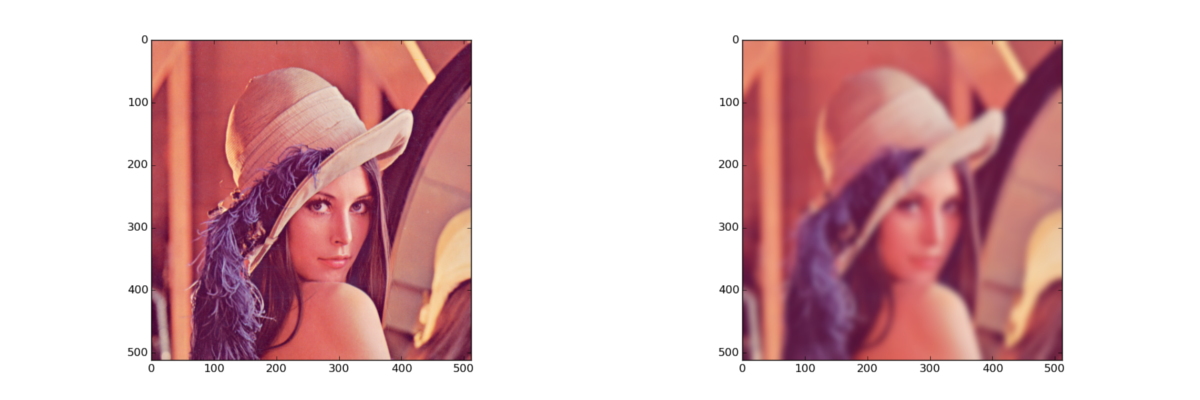
Linear filter: gaussian (3-channel)
import numpy as np import scipy.ndimage as nd from skimage import data, morphology, filter as imfilter import cv2
img = data.lena()
# scipy
dst = np.zeros(img.shape, img.dtype)
for i in xrange(img.shape[2]):
dst[:, :, i] = nd.gaussian_filter(img[:, :, i], 5)
# opencv
dst = cv2.GaussianBlur(img, (0, 0), 5)
# scikit-image
dst = imfilter.gaussian_filter(img, 5, multichannel=True)
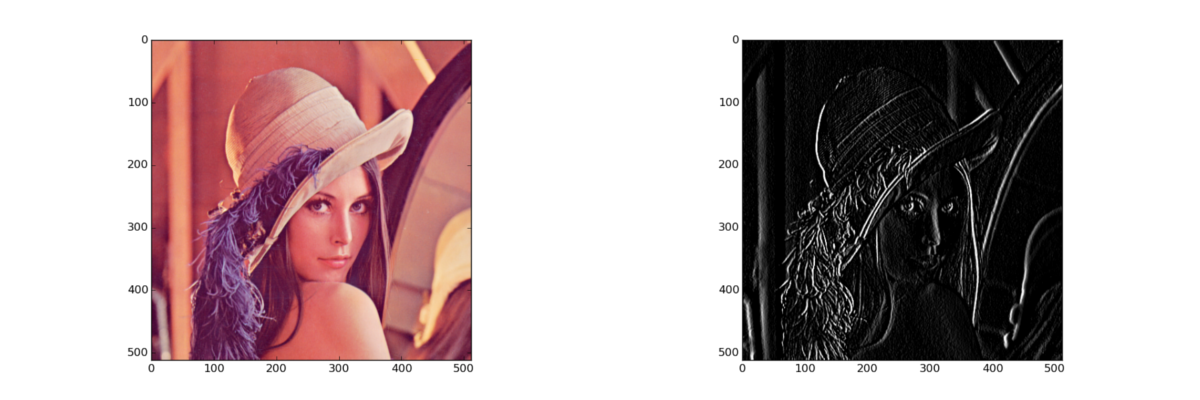
Linear filter: sobel (1-channel)
img = data.lena()[:, :, 1] # scipy dst = nd.sobel(img, 1) # opencv dst = cv2.Sobel(img, cv2.CV_8U, 1, 0) # scikit-image dst = imfilter.hsobel(img)
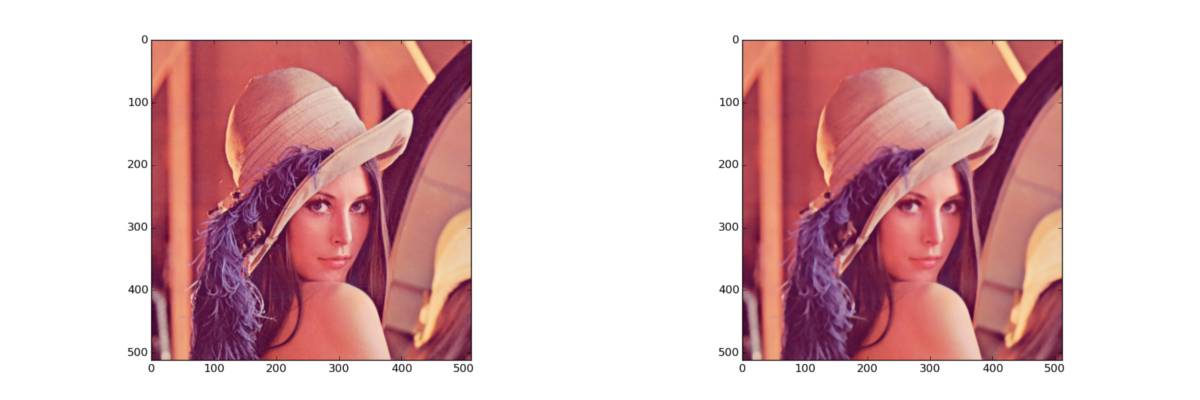
Non-linear filter: median (3-channel)
img = data.lena()
# scipy
dst = np.zeros(img.shape, img.dtype)
for i in xrange(img.shape[2]):
dst[:, :, i] = nd.median_filter(img[:, :, i], 5)
# opencv
dst = cv2.medianBlur(img, 5)
Morphological operation: dilation (1-channel)
img = np.where(data.coins() > 128, 1, 0).astype(np.uint8) kernel = np.ones((5,5), np.uint8) # opencv dst = cv2.dilate(img, kernel, iterations=1) # scipy dst = nd.binary_dilation(img, kernel, iterations=1) # scikit-image dst = morphology.binary_dilation(img, kernel)
Results
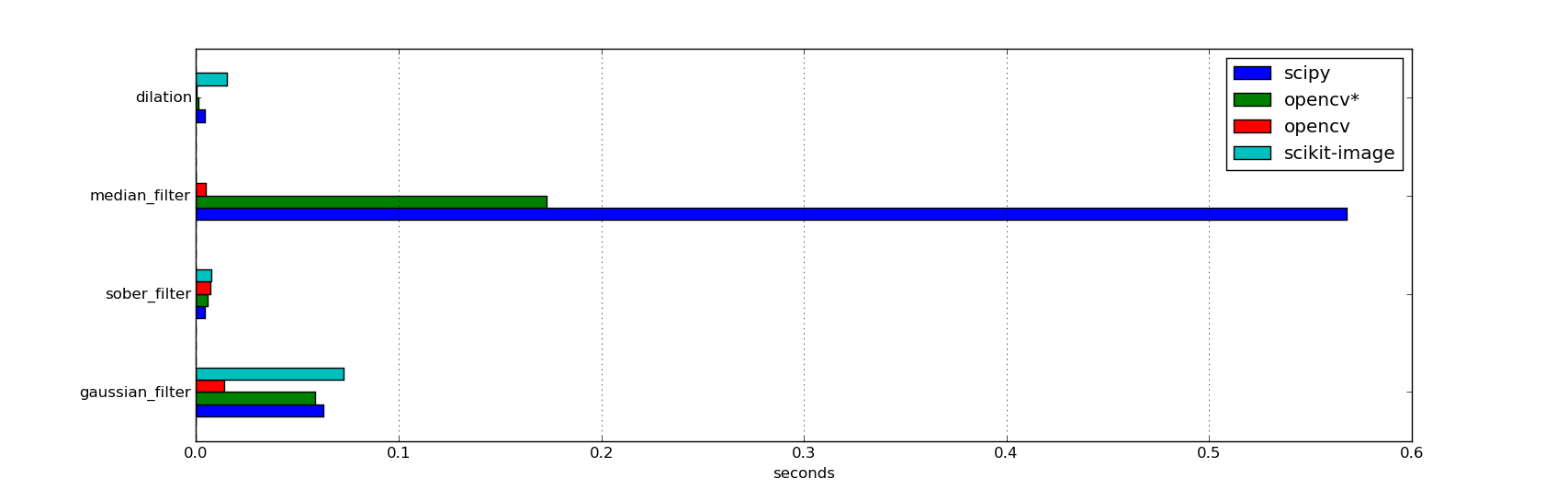
We can see a general better performance in OpenCV (with optimization enabled), specially in a non-linear operation (median filter); a light difference in an easy computable filter as the sobel; and the scikit-image in the last position of performance.
| gaussian filter (rgb) | sobel filter (gray) | median filter (rgb) | dilation (binary) | |
|---|---|---|---|---|
| scipy | 0.0626 | 0.00419 | 0.568 | 0.00452 |
| opencv* | 0.0588 | 0.00592 | 0.173 | 0.00118 |
| opencv | 0.0139 | 0.00684 | 0.0052 | 0.000104 |
| scikit-image | 0.0729 | 0.00746 | 0.0154 | |
|
Time (in seconds) used per process for each of the four libraries (the less the better) * OpenCV with optimization disabled |
||||
import pandas as pd
import matplotlib.pyplot as plt
data = pd.DataFrame(
[[0.0626, 0.00419, 0.568, 0.00452],
[0.0588, 0.00592, 0.173, 0.00118],
[0.0139, 0.00684, 0.005, 0.000104],
[0.0729, 0.00746, np.nan, 0.0154]],
index=['scipy', 'opencv*', 'opencv', 'scikit-image'],
columns=['gaussian_filter', 'sober_filter', 'median_filter', 'dilation'])
data.T.plot(kind='barh', grid=False)
plt.xlabel('seconds')
plt.axes().xaxis.grid(True)
plt.show()